Introduction¶
This is a demo of some of the features implemented in the package PhyloNetwork.
Basic functionality¶
We import the main module and create a phylogenetic network given by its eNewick representation.
Note that branch lengths and metadata are allowed.
[1]:
import phylonetwork as pn
[2]:
net = pn.PhylogeneticNetwork(eNewick='((D[&Theta=0.005]:0.01034,(C[&Theta=0.005]:0.0098)#H1[&Theta=0.005,gamma=0.4051]:4.9969E-4)[&Theta=0.005]:0.04069,(A[&Theta=0.005]:0.0339,(#H1[&Theta=0.005]:0.00489,B[&Theta=0.005]:0.01474)S4[&Theta=0.005]:0.01918)[&Theta=0.005]:0.01710);')
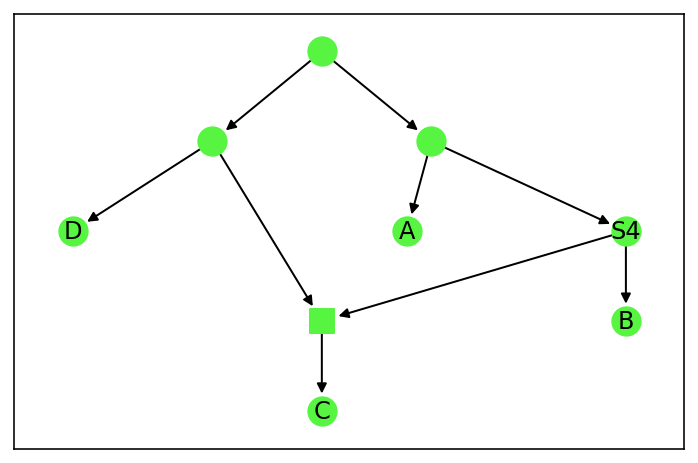
A graphical representation can be given (if the optional packages matplotlib and pygraphviz are available). The labels are shown, and reticulations are indicated by square nodes, while tree nodes are indicated by circle nodes.
[3]:
net.draw()
We obtain some basic information from the network provided by the underlying networkx class
[4]:
net.nodes
[4]:
NodeView(('_1', '_2', '_3', '#1', '_4', '_5', '_6', '_7', '_8'))
[5]:
net.nodes(data=True)
[5]:
NodeDataView({'_1': {}, '_2': {'metadata': {'Theta': '0.005'}}, '_3': {'metadata': {'Theta': '0.005'}, 'label': 'D'}, '#1': {'metadata': {'Theta': '0.005'}}, '_4': {'metadata': {'Theta': '0.005'}, 'label': 'C'}, '_5': {'metadata': {'Theta': '0.005'}}, '_6': {'metadata': {'Theta': '0.005'}, 'label': 'A'}, '_7': {'metadata': {'Theta': '0.005'}, 'label': 'S4'}, '_8': {'metadata': {'Theta': '0.005'}, 'label': 'B'}})
[6]:
net.edges(data=True)
[6]:
OutEdgeDataView([('_1', '_2', {'length': 0.04069}), ('_1', '_5', {'length': 0.0171}), ('_2', '_3', {'length': 0.01034}), ('_2', '#1', {'length': 0.00049969}), ('#1', '_4', {'length': 0.0098}), ('_5', '_6', {'length': 0.0339}), ('_5', '_7', {'length': 0.01918}), ('_7', '#1', {'length': 0.00489}), ('_7', '_8', {'length': 0.01474})])
Some other information is specific to the module
[7]:
net.leaves
[7]:
{'_3', '_4', '_6', '_8'}
[8]:
net.labeling_dict
[8]:
{'_3': 'D', '_4': 'C', '_6': 'A', '_7': 'S4', '_8': 'B'}
[9]:
net.taxa
[9]:
{'A', 'B', 'C', 'D', 'S4'}
[10]:
net.is_tree_child()
[10]:
True
[11]:
net.root
[11]:
'_1'
[12]:
net.mu_dict
[12]:
{'_3': array([0, 0, 0, 1, 0]),
'_4': array([0, 0, 1, 0, 0]),
'#1': array([0, 0, 1, 0, 0]),
'_2': array([0, 0, 1, 1, 0]),
'_6': array([1, 0, 0, 0, 0]),
'_8': array([0, 1, 0, 0, 0]),
'_7': array([0, 1, 1, 0, 1]),
'_5': array([1, 1, 1, 0, 1]),
'_1': array([1, 1, 2, 1, 1])}
[13]:
net.cluster(net.root)
[13]:
{'A', 'B', 'C', 'D', 'S4'}
[14]:
for h in net.reticulations:
print(h, net.cluster(h))
#1 {'C'}
Generation of trees¶
[14]: